












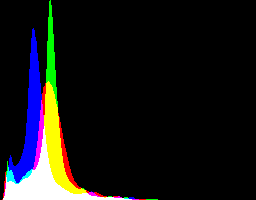

2.96917228 0

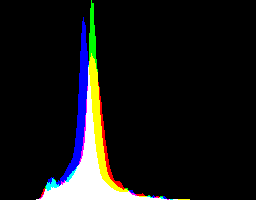

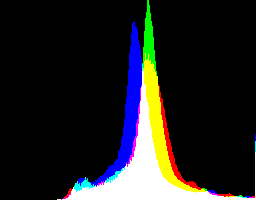

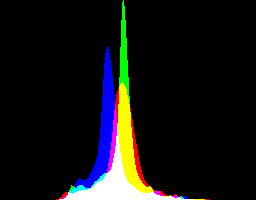

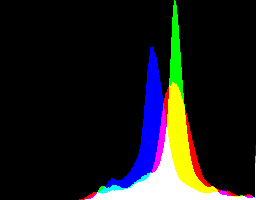
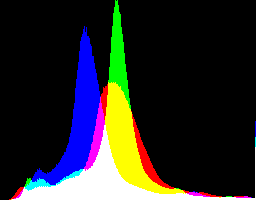
What are the best dcraw gamma and auto-brighten settings for various purposes?
dcraw reads a raw camera file and creates a tiff output. It has a variety of settings. This page considers two: "-g" for gamma, and "-W" which turns off auto-brighten. What are the best settings for two purposes:
For "future processing", the example used in this page is eqLimit.bat. This is a process that makes good first-pass images, and is also representative of manual intervention that may be desired.
Image processing is performed on full-size images. Results are heavily reduced for illustration on the web page.
An ImageMagick bug in version 6.9.1-6 causes "histogram:" images to be of MIFF format, even though they are named ".PNG". See bug report Histogram images. So I use an older version of IM for that task:
set IMH=%IMG691%
The script gameql.bat creates a variety of results: different "-g" settings to dcraw, with or without "-W", before and after eqLimit.bat.
Note that dcraw auto-brightens by default. To turn it off, use -W.
set SRC=%PICTLIB%20130713\AGA_1372.NEF call %PICTBAT%gameql %SRC%
I include the results for -g 1 4.5, though it seems identical to -g 1 0. This is logical; when there is no gamma-shift, a toe-slope is meaningless.
| -g setting | No auto-brighten | Auto-brighten |
|---|---|---|
| 1 0 |


|


|
| 1 4.5 |


|


|
| 2.2222 0 |


|


|
| 2.2222 4.5 |


|


|
| 2.2222 12.92 |


|


|
| no -g setting |


|


|
| gamMaxSd
2.96917228 0 |


|


|
| 4 0 |


|


|
The quantitative data below shows that all the results span the full range from 0.0 to 1.0.
If the purpose is to get an image that is immediately usable, the "auto-brighten" image is superior, whatever the gamma setting. However, auto-brighten has clipped highlights in all cases, and none of the -H settings cure this.
Of the no auto-brighten results, I prefer the one with gamMaxSd.
Of the auto-brighten results, I prefer the ones with the 2.2 gamma.
The images created by dcraw are then processed with eqLimit.bat:
| -g setting | No auto-brighten | Auto-brighten |
|---|---|---|
| 1 0 |

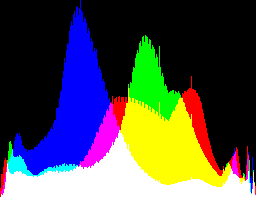
|

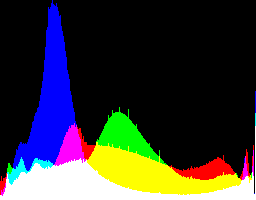
|
| 1 4.5 |

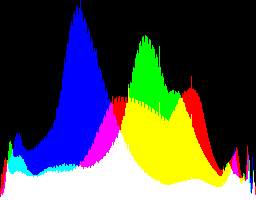
|

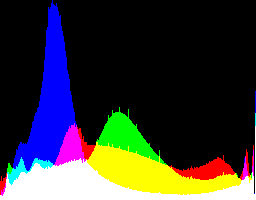
|
| 2.2222 0 |

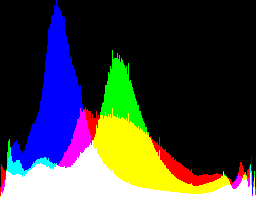
|

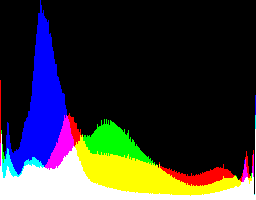
|
| 2.2222 4.5 |

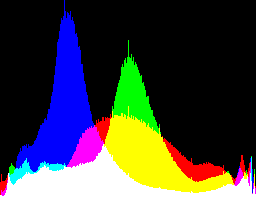
|

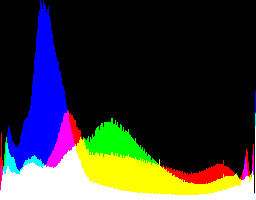
|
| 2.2222 12.92 |

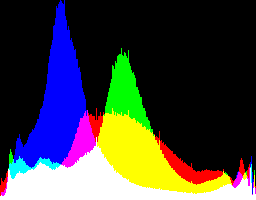
|

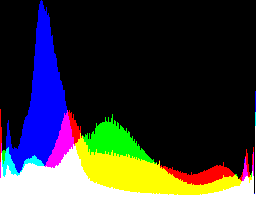
|
| no -g setting |

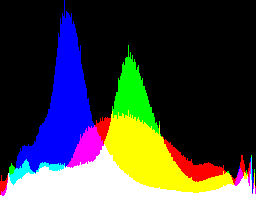
|

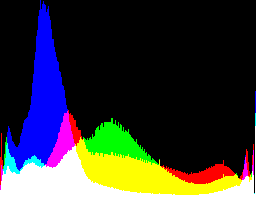
|
| gamMaxSd
2.96917228 0 |


|


|
| 4 0 |

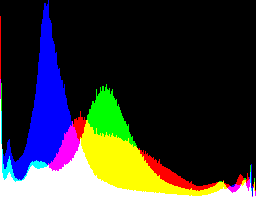
|

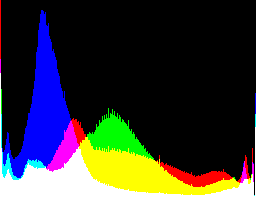
|
Auto-brighten followed by eqLimit creates very high contrast and saturation.
With no auto-brighten, the results are mostly acceptable visually. When the -g gamma increases, the results become darker, and shadows clip at gamMaxSd and 4. Visually, the best result is from -g 1 0.
Any processing that moves values up or down the scale, in integer arithmethic, will lose data. Some values that were different before the operation will become equal after the operation. Once they have become equal, they cannot be separated.
One measure of this is hist_entries in the following listing. It makes a histogram of 65536 buckets, incrementing the count of the buckets corresponding to the R, G and B values. The number shown is the proportion of buckets with at least one entry. The higher this number is, the better.
Examining this data, we discover that:
The data loss is not entirely cumulative. After eqLimit.bat, hist_entries varies from 0.204941 to 0.223282 (no auto-brighten) or 0.174423 to 0.176865 (with auto-brighten). Thus, when eqLimit.bat is used, auto-brighten has a large impact on data loss, but gamma has far less impact.
ge_AGA_1372_gnull_nab.tiff MIN=0 MAX=1 MEAN=0.179174 SD=0.0820161 SatMin=0 SatMax=0.81387 SatMean=0.0701837 SatSD=0.0295917 hist_entries=0.54951477 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.44502 SD=0.22138 SatMin=0 SatMax=1 SatMean=0.224882 SatSD=0.107574 hist_entries=0.057632446 ge_AGA_1372_g1_0_nab.tiff MIN=0 MAX=1 MEAN=0.0532561 SD=0.0412141 SatMin=0 SatMax=0.699748 SatMean=0.0282111 SatSD=0.0190547 hist_entries=0.6098175 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.48936 SD=0.222475 SatMin=0 SatMax=1 SatMean=0.23306 SatSD=0.105319 hist_entries=0.054641724 ge_AGA_1372_g1_4-5_nab.tiff MIN=0 MAX=1 MEAN=0.0532561 SD=0.0412141 SatMin=0 SatMax=0.699748 SatMean=0.0282111 SatSD=0.0190547 hist_entries=0.6098175 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.48936 SD=0.222475 SatMin=0 SatMax=1 SatMean=0.23306 SatSD=0.105319 hist_entries=0.054641724 ge_AGA_1372_g2-2222_0_nab.tiff MIN=0 MAX=1 MEAN=0.252565 SD=0.0760093 SatMin=0 SatMax=0.830686 SatMean=0.0647953 SatSD=0.0266475 hist_entries=0.52577209 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.408083 SD=0.221426 SatMin=0 SatMax=1 SatMean=0.219291 SatSD=0.107809 hist_entries=0.061584473 ge_AGA_1372_g2-2222_4-5_nab.tiff MIN=0 MAX=1 MEAN=0.179173 SD=0.0820157 SatMin=0 SatMax=0.81387 SatMean=0.0701833 SatSD=0.0295915 hist_entries=0.54954529 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.444951 SD=0.22135 SatMin=0 SatMax=1 SatMean=0.224846 SatSD=0.107553 hist_entries=0.057693481 ge_AGA_1372_g2-2222_12-92_nab.tiff MIN=0 MAX=1 MEAN=0.224367 SD=0.0788761 SatMin=0 SatMax=0.824292 SatMean=0.067239 SatSD=0.0276506 hist_entries=0.53512573 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.421873 SD=0.22093 SatMin=0 SatMax=1 SatMean=0.221016 SatSD=0.107825 hist_entries=0.059906006 ge_AGA_1372_g2-98764047_0_nab.tiff MIN=0 MAX=1 MEAN=0.356109 SD=0.0791845 SatMin=0 SatMax=0.871122 SatMean=0.0688918 SatSD=0.0271509 hist_entries=0.47833252 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.380472 SD=0.223898 SatMin=0 SatMax=1 SatMean=0.217618 SatSD=0.110419 hist_entries=0.062988281 ge_AGA_1372_g4_0_nab.tiff MIN=0 MAX=1 MEAN=0.460147 SD=0.0765078 SatMin=0 SatMax=0.902083 SatMean=0.0671387 SatSD=0.0262795 hist_entries=0.42617798 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.367158 SD=0.223582 SatMin=0 SatMax=1 SatMean=0.215116 SatSD=0.110027 hist_entries=0.063262939 ge_AGA_1372_gnull_ab.tiff MIN=0 MAX=1 MEAN=0.422196 SD=0.152667 SatMin=0 SatMax=1 SatMean=0.13268 SatSD=0.0554021 hist_entries=0.24537659 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.390264 SD=0.253736 SatMin=0 SatMax=1 SatMean=0.253986 SatSD=0.153644 hist_entries=0.066513062 ge_AGA_1372_g1_0_ab.tiff MIN=0 MAX=1 MEAN=0.214716 SD=0.152318 SatMin=0 SatMax=1 SatMean=0.111801 SatSD=0.0702119 hist_entries=0.24537659 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.421451 SD=0.246159 SatMin=0 SatMax=1 SatMean=0.252058 SatSD=0.145338 hist_entries=0.061935425 ge_AGA_1372_g1_4-5_ab.tiff MIN=0 MAX=1 MEAN=0.214716 SD=0.152318 SatMin=0 SatMax=1 SatMean=0.111801 SatSD=0.0702119 hist_entries=0.24537659 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.421451 SD=0.246159 SatMin=0 SatMax=1 SatMean=0.252058 SatSD=0.145338 hist_entries=0.061935425 ge_AGA_1372_g2-2222_0_ab.tiff MIN=0 MAX=1 MEAN=0.474378 SD=0.138897 SatMin=0 SatMax=1 SatMean=0.120715 SatSD=0.0503922 hist_entries=0.24537659 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.384956 SD=0.252736 SatMin=0 SatMax=1 SatMean=0.251187 SatSD=0.150289 hist_entries=0.065826416 ge_AGA_1372_g2-2222_4-5_ab.tiff MIN=0 MAX=1 MEAN=0.422195 SD=0.152667 SatMin=0 SatMax=1 SatMean=0.13268 SatSD=0.0554022 hist_entries=0.24537659 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.390262 SD=0.253736 SatMin=0 SatMax=1 SatMean=0.253985 SatSD=0.153645 hist_entries=0.066619873 ge_AGA_1372_g2-2222_12-92_ab.tiff MIN=0 MAX=1 MEAN=0.454548 SD=0.144136 SatMin=0 SatMax=1 SatMean=0.125269 SatSD=0.0522913 hist_entries=0.24537659 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.386865 SD=0.253125 SatMin=0 SatMax=1 SatMean=0.252205 SatSD=0.15157 hist_entries=0.066223145 ge_AGA_1372_g2-98764047_0_ab.tiff MIN=0 MAX=1 MEAN=0.569266 SD=0.124021 SatMin=0 SatMax=1 SatMean=0.109377 SatSD=0.0440367 hist_entries=0.24537659 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.384677 SD=0.25076 SatMin=0 SatMax=1 SatMean=0.249194 SatSD=0.145433 hist_entries=0.06539917 ge_AGA_1372_g4_0_ab.tiff MIN=0 MAX=1 MEAN=0.653312 SD=0.106873 SatMin=0 SatMax=1 SatMean=0.0947514 SatSD=0.0379474 hist_entries=0.24537659 Result of eqLimit.bat: MIN=0 MAX=1 MEAN=0.387941 SD=0.247856 SatMin=0 SatMax=1 SatMean=0.247241 SatSD=0.139171 hist_entries=0.063949585
A point of interest here: when the toe-slope is zero, the highest standard deviation comes from gamMaxSd, of course. But the setting -g 2.2222 4.5 gives a slightly higher SD. This suggests a future avenue to explore: varying the toe-slope for the maximum SD.
If we want an sRGB version, we have the choice of making it with dcraw or IM. The dcraw version comes directly from dcraw, with no options for gamma or -W, so it is auto-brightened. The IM version comes from "-set colorspace RGB -colorspace sRGB", from the dcraw output of gamma 1, with auto-brighten.
The numbers from IM's sRGB image are:
MIN=0 MAX=1 MEAN=0.472245 SD=0.142847 SatMin=0 SatMax=1 SatMean=0.124783 SatSD=0.0513867 hist_entries=0.24537659
They are close to the numbers from dcraw's sRGB image. The IM version has a higher mean and lower SD. IM's saturation has a higher maximum, lower mean and lower SD. The number of hist_entries is identical.
The images are:
| dcraw's sRGB |


|
| IM's sRGB |

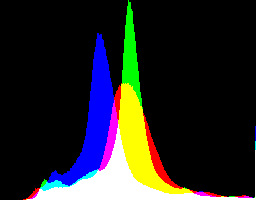
|
Confirming the numbers: IM's version has the histogram pushed right slightly, lightening the entire image, especially the shadows, and reducing most contrast.
I see no obvious preference for either version, but standardising on one is useful. I choose to use dcraw's version.
Both sRGB images above have clipped highlights, caused by dcraw's auto-brighten. An alternative is to use a non-auto-brightened source, given a contrast-limited equalisation, further tamed by "+sigmoidal-contrast" to push darkish and lightish tones towards the middle, which reduces the standard deviation and the saturation. The result isn't photometrically sRGB, but it is aesthetically pleasing, useful for further processing, and is not clipped. Here are two versions: the first from dcraw "-g 1 0", the second from unspecified "-g". They are slightly different, but I don't have a clear preference.
%IMH%convert ^ ge_%RAW_BASE%_g1_0_nab_eql.tiff ^ +sigmoidal-contrast 10,50%% ^ +write ge_eql_sRGB10.tiff ^ -define histogram:unique-colors=false ^ +write histogram:ge_eql_sRGB10_hist.png ^ %WEB_SIZE% ^ ge_eql_sRGB10_sm.jpg |

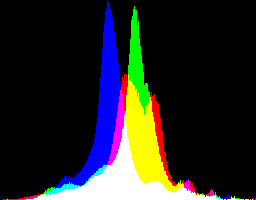
|
%IMH%convert ^ ge_%RAW_BASE%_gnull_nab_eql.tiff ^ +sigmoidal-contrast 10,50%% ^ +write ge_eql_sRGB.tiff ^ -define histogram:unique-colors=false ^ +write histogram:ge_eql_sRGB_hist.png ^ %WEB_SIZE% ^ ge_eql_sRGB_sm.jpg |

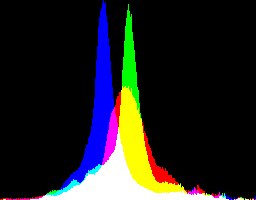
|
We may prefer an sRGB version that has a specified gamma. What gamma?
|
Make gamma 0.5 in RGB colorspace, which is about 0.73 in sRGB space. %IMH%convert ^ ge_%RAW_BASE%_g1_0_ab.tiff ^ -auto-level ^ -auto-gamma ^ -set colorspace RGB -colorspace sRGB ^ +write ge_alg_sRGB.tiff ^ -define histogram:unique-colors=false ^ +write histogram:ge_alg_sRGB_hist.png ^ %WEB_SIZE% ^ ge_alg_sRGB_sm.jpg |

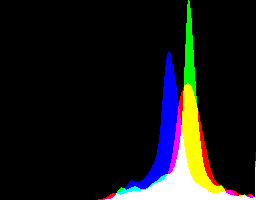
|
|
Make gamma 0.5 in sRGB colorspace. %IMH%convert ^ ge_%RAW_BASE%_g1_0_ab.tiff ^ -set colorspace RGB -colorspace sRGB ^ -auto-level ^ -auto-gamma ^ +write ge_alg_sRGB.tiff ^ -define histogram:unique-colors=false ^ +write histogram:ge_sRGB_alg_hist.png ^ %WEB_SIZE% ^ ge_sRGB_alg_sm.jpg |

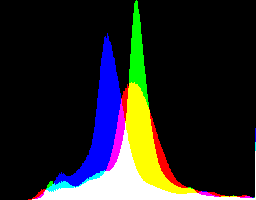
|
The questions were:
What are the best gamma and auto-brightness settings for two purposes:
My answers are:
I also create an _eql version. Thus, we have three simple commands:
dcraw -v -H 0 -6 -w -o 1 -T -O out_sRGB.tiff in.nef dcraw -v -H 0 -6 -w -W -g 1 0 -o 0 -T -O out_raw.tiff in.nef call %PICTBAT%eqLimit out_raw.tiff . . . out_eql.tiff
The file out_raw.tiff is visually horrible (very dark), but useful for further processing. The two visual outputs are out_sRGB.tiff and out_eql.tiff. Comparing these outputs to in-camera JPEGs made by a Nikon D800, I find that:
The dcraw commands shown on this page use -w to use camera white balance, if possible, and -H 0 which uses camera white balance even if it causes clipping. I do this for convenience. In practise, I do not recommend permitting dcraw to clip images. A better solution is either to use one of the other -H XX options, or to extract the white-balance parameters and maximum pixel values, then re-calculate the parameters such that no clipping occurs. This is slow because dcraw must be called twice.
We don't need to keep the large TIFF files, so delete them:
del ge_*_*.tiff
For convenience, .bat scripts are also available in a single zip file. See Zipped BAT files.
rem Tests dcraw and eqLimit with a variety of parameters.
rem %1 is input raw file.
if "%IMH%"=="" set IMH=%IM%
set IN_RAW=%1
if not exist "%1" (
echo %0: "%1" does not exist
exit /B 1
)
set RAW_BASE=%~n1
set COUNT_PROC=-separate +append -process 'mkhisto norm' -fill White +opaque Black
set WEB_SIZE=-resize 300x300
set OUT_LIS=ge_gameql.lis
del %OUT_LIS% 2>nul
set TMP_SRGB=%TEMP%\ge_IM_srgb.tiff
set OUT_TIFF=%TEMP%\ge_temp.tiff
%IMG7%dcraw -v -6 -w -W -g 1 0 -o 0 -T -O %OUT_TIFF% %IN_RAW%
call %PICTBAT%gamMaxSd %OUT_TIFF% 0.05 10
call :geOne %IN_RAW% 0 . .
call :geOne %IN_RAW% 0 1 0
call :geOne %IN_RAW% 0 1 4.5
call :geOne %IN_RAW% 0 2.2222 0
call :geOne %IN_RAW% 0 2.2222 4.5
call :geOne %IN_RAW% 0 2.2222 12.92
call :geOne %IN_RAW% 0 %gmsGAM_MAX_SD% 0
call :geOne %IN_RAW% 0 4 0
call :geOne %IN_RAW% 1 . .
call :geOne %IN_RAW% 1 1 0
call :geOne %IN_RAW% 1 1 4.5
call :geOne %IN_RAW% 1 2.2222 0
call :geOne %IN_RAW% 1 2.2222 4.5
call :geOne %IN_RAW% 1 2.2222 12.92
call :geOne %IN_RAW% 1 %gmsGAM_MAX_SD% 0
call :geOne %IN_RAW% 1 4 0
rem ===============================
rem
rem Try sRGB version from IM.
%IMG7%magick ^
ge_%RAW_BASE%_g1_0_ab.tiff ^
-set colorspace RGB -colorspace sRGB ^
%TMP_SRGB%
%IMG7%magick ^
%TMP_SRGB% ^
-format "MIN=%%[fx:minima] MAX=%%[fx:maxima] MEAN=%%[fx:mean] SD=%%[fx:standard_deviation]\n" ^
-write info: ^
^( +clone ^
-colorspace HCL ^
-format "SatMin=%%[fx:minima.g] SatMax=%%[fx:maxima.g] SatMean=%%[fx:mean.g] SatSD=%%[fx:standard_deviation.g]\n" ^
-write info: ^
+delete ^
^) ^
NULL:>ge_im_srgb.lis
if ERRORLEVEL 1 exit /B 1
%IM7DEV%magick ^
%TMP_SRGB% ^
%COUNT_PROC% ^
-format "hist_entries=%%[fx:mean]\n" ^
info: >>ge_im_srgb.lis
if ERRORLEVEL 1 exit /B 1
%IMG7%magick ^
%TMP_SRGB% ^
-define histogram:unique-colors=false ^
+write histogram:ge_im_srgb_hist.png ^
%WEB_SIZE% ^
ge_im_srgb_sm.jpg
if ERRORLEVEL 1 exit /B 1
exit /B 0
rem ============ Subroutines ===================
rem
rem Params to geOne:
rem %1 raw filename
rem %2 0 if -W (no auto-brighten); 1 if auto-brighten
rem %3 first -g parameter (or "." for no -g)
rem %4 second -g parameter
:geOne
set G_ARG=
set G_NAME=gnull
set G1=%3
set G2=%4
if not "%3"=="." (
set G_ARG=-g %3 %4
set G_NAME=g%G1:.=-%_%G2:.=-%
)
echo G_NAME=%G_NAME%
set W_ARG=
set AB_NAME=ab
if "%2"=="0" (
set W_ARG=-W
set AB_NAME=nab
)
set BASE=ge_%RAW_BASE%_%G_NAME%_%AB_NAME%
set OUT_TIFF=%BASE%.tiff
set OUT_EQL=%BASE%_eql.tiff
rem set OUT_LIS=%BASE%.lis
rem del %OUT_LIS% 2>nul
echo OUT_TIFF=%OUT_TIFF%
%IMG7%dcraw -v -H 0 -6 -w %W_ARG% %G_ARG% -o 0 -T -O %OUT_TIFF% %1
if ERRORLEVEL 1 exit /B 1
%IMG7%magick ^
%OUT_TIFF% ^
-format "%%f\nMIN=%%[fx:minima] MAX=%%[fx:maxima] MEAN=%%[fx:mean] SD=%%[fx:standard_deviation]\n" ^
-write info: ^
^( +clone ^
-colorspace HCL ^
-format "SatMin=%%[fx:minima.g] SatMax=%%[fx:maxima.g] SatMean=%%[fx:mean.g] SatSD=%%[fx:standard_deviation.g]\n" ^
-write info: ^
+delete ^
^) ^
NULL:>>%OUT_LIS%
if ERRORLEVEL 1 exit /B 1
%IM7DEV%magick ^
%OUT_TIFF% ^
%COUNT_PROC% ^
-format "hist_entries=%%[fx:mean]\n\n" ^
info: >>%OUT_LIS%
if ERRORLEVEL 1 exit /B 1
call %PICTBAT%eqLimit %OUT_TIFF%
if ERRORLEVEL 1 exit /B 1
echo Result of eqLimit.bat: >>%OUT_LIS%
%IMG7%magick ^
%OUT_EQL% ^
-format "MIN=%%[fx:minima] MAX=%%[fx:maxima] MEAN=%%[fx:mean] SD=%%[fx:standard_deviation]\n" ^
-write info: ^
^( +clone ^
-colorspace HCL ^
-format "SatMin=%%[fx:minima.g] SatMax=%%[fx:maxima.g] SatMean=%%[fx:mean.g] SatSD=%%[fx:standard_deviation.g]\n" ^
-write info: ^
+delete ^
^) ^
NULL:>>%OUT_LIS%
%IM7DEV%magick ^
%OUT_EQL% ^
%COUNT_PROC% ^
-format "hist_entries=%%[fx:mean]\n\n\n\n" ^
info: >>%OUT_LIS%
if ERRORLEVEL 1 exit /B 1
%IMG7%magick ^
%OUT_TIFF% ^
-define histogram:unique-colors=false ^
+write histogram:%BASE%_hist.png ^
%WEB_SIZE% ^
%BASE%_sm.jpg
if ERRORLEVEL 1 exit /B 1
%IMG7%magick ^
%OUT_EQL% ^
-define histogram:unique-colors=false ^
+write histogram:%BASE%_eql_hist.png ^
%WEB_SIZE% ^
%BASE%_eql_sm.jpg
if ERRORLEVEL 1 exit /B 1
rem
rem ============ End Subroutines ===================
All images on this page were created by the commands shown, using:
%IMG7%magick -version
Version: ImageMagick 7.1.0-42 Q16-HDRI x64 396d87c:20220709 https://imagemagick.org Copyright: (C) 1999 ImageMagick Studio LLC License: https://imagemagick.org/script/license.php Features: Cipher DPC HDRI OpenCL Delegates (built-in): bzlib cairo freetype gslib heic jng jp2 jpeg jxl lcms lqr lzma openexr pangocairo png ps raqm raw rsvg tiff webp xml zip zlib Compiler: Visual Studio 2022 (193231332)
Source file for this web page is gameql.h1. To re-create this web page, execute "procH1 gameql".
This page, including the images, is my copyright. Anyone is permitted to use or adapt any of the code, scripts or images for any purpose, including commercial use.
Anyone is permitted to re-publish this page, but only for non-commercial use.
Anyone is permitted to link to this page, including for commercial use.
Page version v1.0 24-Dec-2014.
Page created 03-Sep-2022 04:33:33.
Copyright © 2022 Alan Gibson.